DEGs: What are Differentially Expressed Genes?
Differentially Expressed Genes (DEGs)
Differentially Expressed Genes (DEGs) refer to genes whose expression levels significantly increase or decrease between different conditions or groups. After comprehensive gene expression analyses such as RNA-Seq or microarrays, differential expression analysis is commonly performed to identify differentially expressed genes (DEGs).
For example, when conducting RNA-Seq analysis on humans, the expression levels of tens of thousands of genes are obtained. Interpreting these results directly can be challenging. By extracting genes whose expression levels have significantly increased or decreased from among these tens of thousands, biological interpretation becomes more manageable.
Various software has been developed for the identification of differentially expressed genes. Well-known examples include edgeR, DESeq2, and Ballgown.
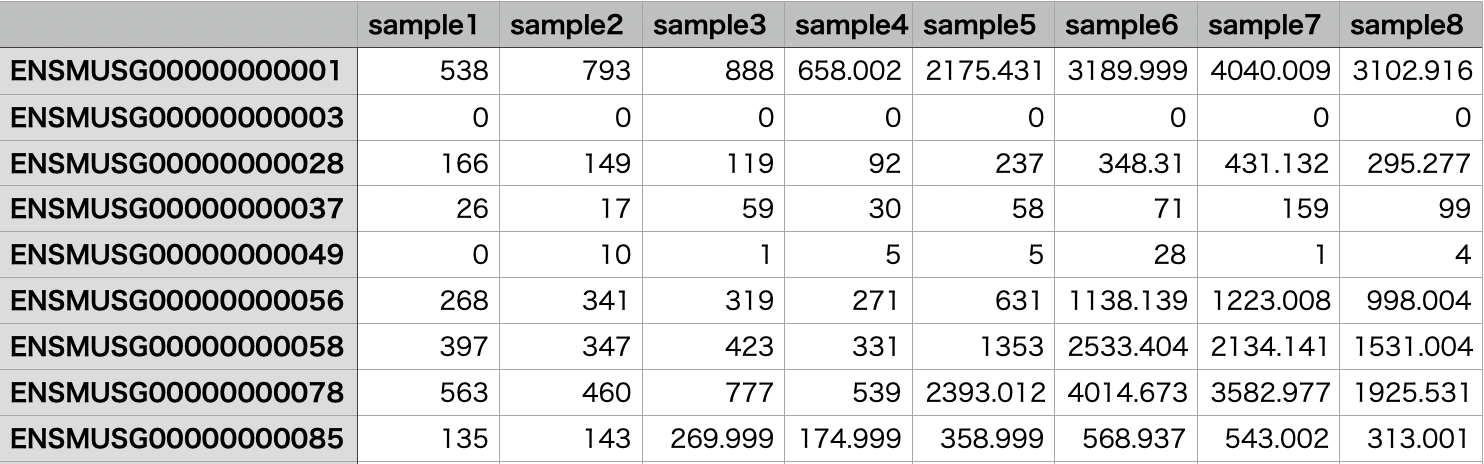
Example of an Expression Level Table
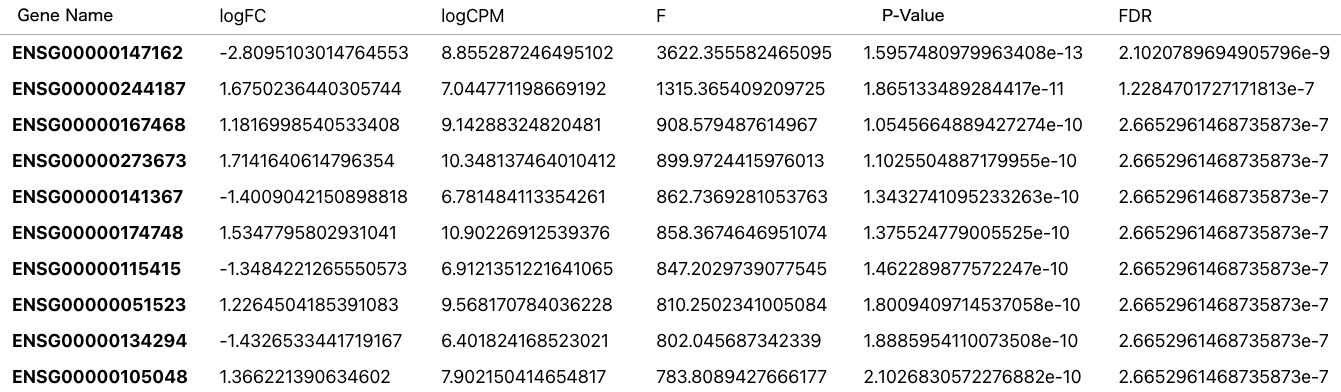
Example of Differentially Expressed Genes
Functional Analysis After Identification of Differentially Expressed Genes
Identification of differentially expressed genes yields a list of genes. When the list contains only a few genes, it is possible to examine each one individually. However, interpretation can be challenging when the list includes a large number of genes.
Therefore, functional analysis is often conducted after the identification of differentially expressed genes. This analysis examines which functions are predominantly associated with the genes listed.
In functional analysis, it is common to focus on Gene Ontology or pathways. These are often referred to as gene ontology analysis and pathway analysis, respectively.
Please see below for more details on gene ontology analysis.
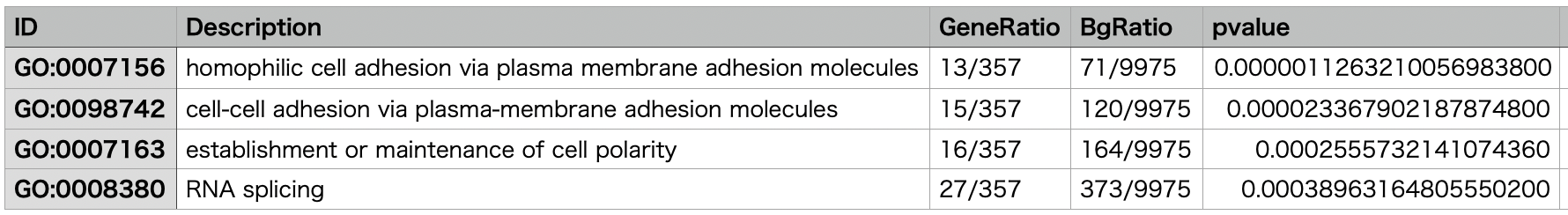
Example of Gene Ontology Analysis
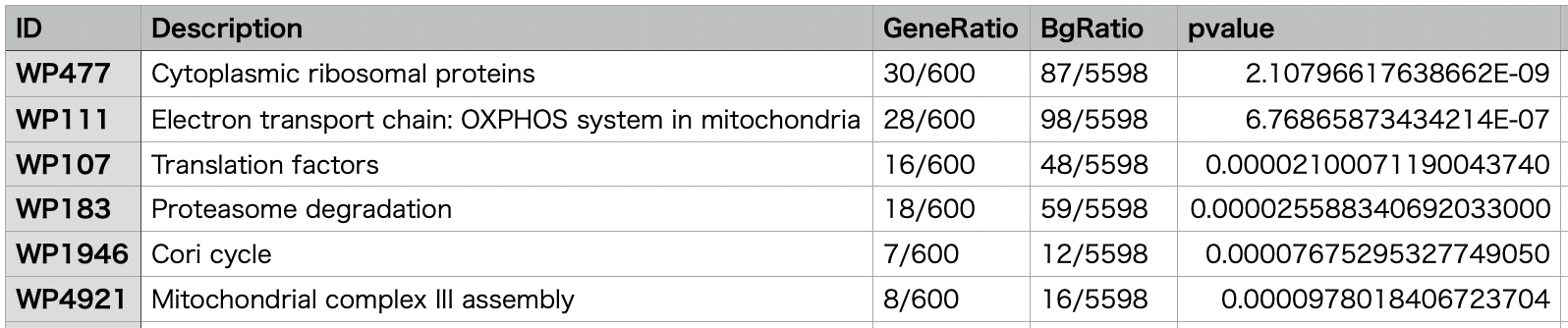
Example of Pathway Analysis
RNA-Seq Data Analysis Software
This is an RNA-Seq Data Analysis Software recommended for those who:
✔︎ Seeking to avoid outsourcing or collaboration for RNA-Seq data analysis.
✔︎ Lacking time to learn RNA-Seq data analysis.
✔︎ Frustrated by the complexity of existing tools.
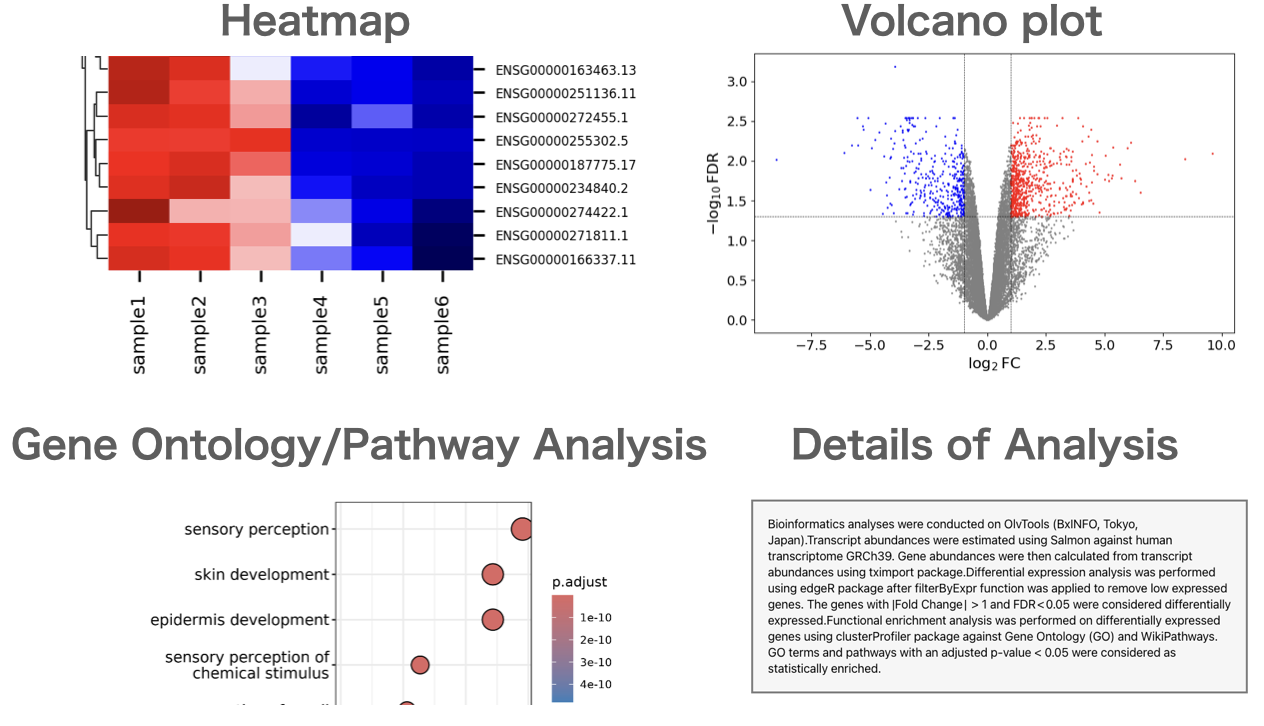
Users can perform gene expression quantification, identification of differentially expressed genes, gene ontology(GO) analysis, pathway analysis, as well as drawing volcano plots, MA plots, and heatmaps.
About the Author
BxINFO LLC
A research support company specializing in bioinformatics.
We provide tools and information to support life science research, with a focus on RNA-Seq analysis.