Free Primer Design Tool
Enter a template sequence and click the "Design Primers" button to generate the optimal primer pair.
About Primer Design
Proper primer design is crucial for performing PCR and Sanger sequencing successfully. When designing primers, you need to pay attention to the following points:
- Length of the primer
- Length of the amplicon
- GC content of the primer
- Tm value of the primer
- Absence of secondary structure in the primer
- Specificity of the primer
Finding sequences that meet these criteria manually is extremely difficult. This tool automates the search for suitable sequences, making primer design more efficient.
How to Use
- Enter a template sequence and click the "Design Primers" button to generate the optimal primer pair.
- After entering the template sequence, click "Set" in the "Target Region" section to specify the region to be included in the amplicon.
- After entering the template sequence, click "Set" in the "Overlap" section to design primers that overlap a specified region.
- After entering the template sequence, click "Set" in the "Excluded Region" section to specify regions to be excluded from primer design.
- In the advanced settings, you can specify the primer length, amplicon length, GC content, and Tm value.
- After primer design, click "View on Template Sequence" to check the positions of primers, target regions, and excluded regions.
References
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M and Rozen SG. Primer3--new capabilities and interfaces. Nucleic Acids Res. 2012 Aug 1;40(15):e115.
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. (1990) “Basic local alignment search tool.” J. Mol. Biol. 215:403-410.
RNA-Seq Data Analysis Software
This is an RNA-Seq Data Analysis Software recommended for those who:
✔︎ Seeking to avoid outsourcing or collaboration for RNA-Seq data analysis.
✔︎ Lacking time to learn RNA-Seq data analysis.
✔︎ Frustrated by the complexity of existing tools.
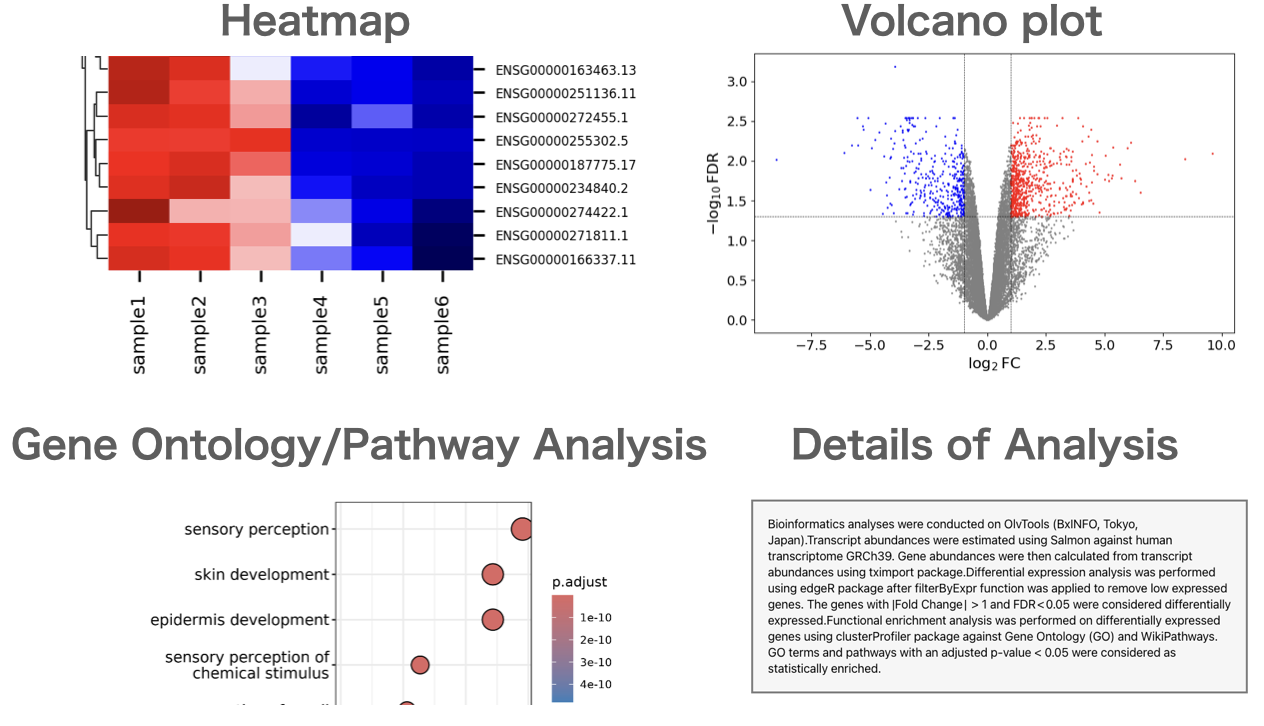
Users can perform gene expression quantification, identification of differentially expressed genes, gene ontology(GO) analysis, pathway analysis, as well as drawing volcano plots, MA plots, and heatmaps.